A team of bioengineers has successfully utilized AI to resurrect molecules from our ancient past.
This intriguing study utilized machine learning (ML) to analyze protein data from modern humans (Homo sapiens) as well as from our extinct relatives, Neanderthals (Homo neanderthalensis) and Denisovans, an extinct species or subspecies of human from Asia.
The study discovered molecules with the potential to combat disease-causing bacteria. These techniques, dubbed ‘de-extinction,’ could potentially serve as the basis for locating new antibiotics.
“We’re motivated by the notion of bringing back molecules from the past to address problems that we have today,” Cesar de la Fuente, study co-author and bioengineer at the University of Pennsylvania in Philadelphia, told Nature.
If this makes you think of a certain dinosaur-based film franchise – that’s precisely what inspired the research team.
De la Fuente said, “We started actually thinking about Jurassic Park,” he says. Rather than bringing dinosaurs back to life, as scientists did in the 1993 film, the team came up with a more feasible idea: “Why not bring back molecules?”
There is immense pressure on researchers to locate new antibiotics, with few effective drugs discovered in the last 30 years.
Meanwhile, the prevalence of antibiotic-resistant bacteria is increasing, so novel treatments are welcomed. AI provides numerous avenues to explore those novel treatments, with a range of studies leveraging the technology to uncover hidden and unknown clinically relevant molecules.
Many organisms generate short protein subunits known as peptides with antimicrobial properties. Several antimicrobial peptides, predominantly sourced from bacteria, are used to treat infections.
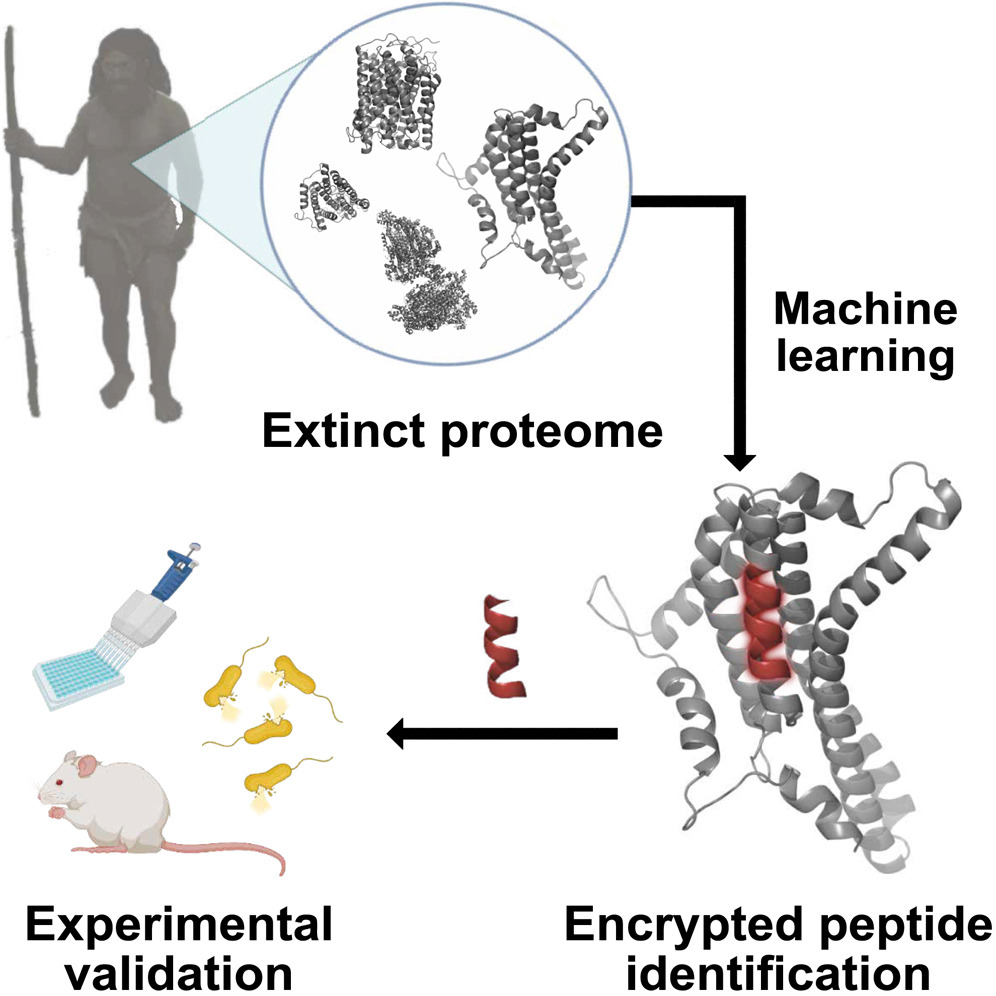
The proteins of extinct species – including humans – could serve as a valuable yet underexplored resource for developing new antibiotics.
The researchers trained an ML algorithm to identify locations on human proteins where they are sliced into peptides. They applied this algorithm to publicly available protein sequences from H. sapiens, H. neanderthalensis, and Denisovans, identifying new peptides based on the characteristics of known antimicrobial peptides.
It takes only a few weeks to identify and test potential drug candidates using AI, compared to the traditional process that takes anywhere from 3 to six 6 to discover a new antibiotic.
The team tested a variety of peptides in a laboratory setting to explore their ability to eliminate bacteria. They selected 6 potent peptides — 4 from H. sapiens, 1 from H. neanderthalensis, and 1 from Denisovans. The peptides were tested on mice infected with Acinetobacter baumannii.
Although none of the peptides fully killed the bacteria, they all managed to inhibit the growth of A. baumannii in thigh muscle tissue. 5 molecules killed bacteria in skin abscesses, though high dosages were required.
“Even though the algorithm that we used didn’t yield amazing molecules, I think the concept and the framework represents an entirely new avenue for thinking about drug discovery,” de la Fuente states.
Euan Ashley, a genomics and precision-health expert at Stanford University, commented, “De la Fuente and his colleagues persuaded me that diving into the archaic human genome was an interesting and potentially useful approach.”
How the study worked: a breakdown
- The researchers set out to resurrect (or “de-extinct”) bioactive molecules no longer produced by living organisms. These molecules could potentially be used for drug discovery.
- They used a machine learning (ML) model known as panCleave. This is a “random forest” model, a type of AI algorithm that predicts outcomes based on many inputs. The panCleave model predicted where proteins would be split into smaller parts known as peptides. These peptides sometimes have antimicrobial properties.
- The outcome: The researchers found several peptides (both from modern humans and extinct human relatives) that showed potential as antimicrobial agents when tested in the lab.
- Testing included resistance to being broken by other proteins and the ability to permeate cell membranes. Some of the peptides effectively fought a common bacterial infection in mice’s skin abscesses.
- Machine-learning methods can identify promising peptides for the development of new antibiotics. Moreover, “molecular de-extinction” (resurrecting molecules from extinct organisms) is proposed as a new framework for drug discovery, specifically for finding new antibiotics.
AI’s potential for discovering new drugs at scale is proving immense, and not just antibiotics, but anti-aging drugs and other compounds too.





